Package chemosensors.
Usage
loadUNIMANdata(dataset) subClasses(class) ConcUnitsNames() defaultDataModel() defaultConcUnitsInt() defaultConcUnits() defaultConcUnitsSorption() defaultSet() defaultDataPackage() defaultDataSensorModel() defaultDataDistr() defaultDataSensorNoiseModel() defaultDataSorptionModel() defaultDataDriftNoiseModel()
Arguments
- dataset
- Name of dataset to be loaded.
- class
- Class name.
Value
Character vector of sub-classes.
Character vector of units names.
Description
A tool to set up synthetic experiments in machine olfaction.
Function loadUNIMANdata.
Support function to get sub-classes.
Function to get available names for concentration units.
Function defaultDataModel.
Function defaultConcUnitsInt.
Function defaultConcUnits.
Function defaultConcUnitsSorption.
Function defaultSet
Function defaultDataPackage.
Function defaultDataSensorModel.
Function defaultDataDistr.
Function defaultDataSensorNoiseModel.
Function defaultDataSorptionModel.
Function defaultDataDriftNoiseModel.
Examples
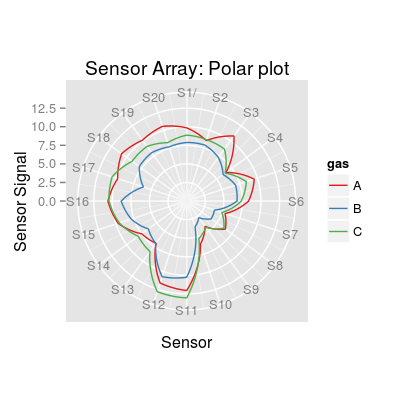
# concentration matrix of 3 gas classes: A, C and AC conc <- matrix(0, 60, 3) conc[1:10, 1] <- 0.01 # A0.01 conc[11:20, 3] <- 0.1 # C0.1 conc[21:30, 1] <- 0.05 # A0.05 conc[31:40, 3] <- 1 # C1 conc[41:50, 1] <- 0.01 # A0.01C0.1 conc[41:50, 3] <- 0.1 # A0.01C0.1 conc[51:60, 1] <- 0.05 # A0.05C1 conc[51:60, 3] <- 1 # A0.05C1 conc <- conc[sample(1:nrow(conc)), ] # sensor array of 20 sensors with parametrized noise parameters sa <- SensorArray(nsensors=20, csd=0.1, ssd=0.1, dsd=0.1) # get information about the array print(sa)SensorArray - enableSorption: TRUE (1) Sensor Model - num 1, 2, 3 ... 3 - beta 2 - 3 gases A, B, C - (first) data model - method: ispline (type: spline) - sensor model: coeffNonneg TRUE -- coefficients (first): 3.4868, 3.4731, 4.3785 ... 4.5575 (2) Sorption Model - knum 1, 2, 3 ... 3 - 3 gases A, B, C (3) Concentration Noise Model - 3 gases A, B, C - csd: 0.1 - noise type: logconc - log-factor: 1, 1, 2 (4) Sensor Noise Model - num 1, 2, 3 ... 3 - 3 gases A, B, C - ssd: 0.1 - noise type: randomWalk - noise-factor: 1, 1, 1, 1, 1, 1, 1, 1, 1 (5) Drift Noise Model - num 1, 2, 3 ... 3 drift common model - method: cpc - ndcomp: 1plot(sa)
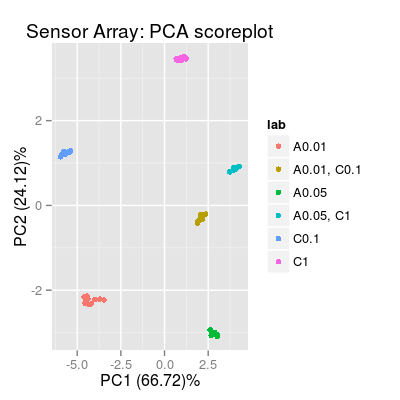
# generate the data sdata <- predict(sa, conc) # plot the data plot(sa, "prediction", conc=conc, sdata=sdata, leg="top")