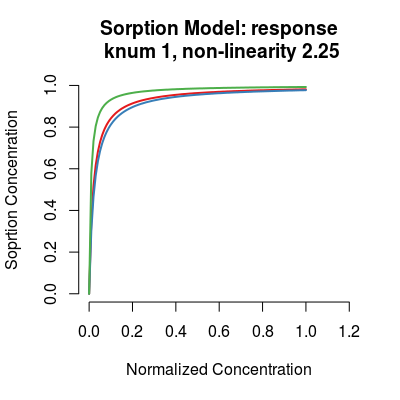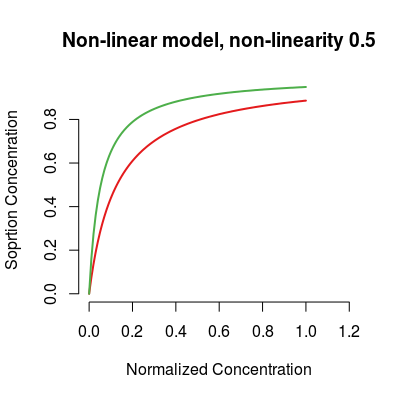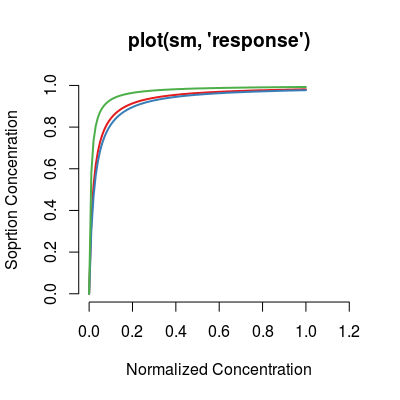Method knum.
Usage
defaultSorptionModel() SorptionModel(...)
Arguments
- ...
- parameters of constructor.
Value
List of the default parameters.
Description
Method knum.
Method concUnitsSorption.
Class SorptionModel controls the the amount
of gas absorbed by the sensor, that emulates the
non-linearity nature intrinsic for the polymeric sensors.
Function to get default constructor parameters of class
SorptionModel.
Constructor method of SorptionModel Class.
Wrapper function SorptionModel.
Details
The model is based on the extended Langmuir isotherm for
a multi-component gas mixture, and has two parameters per
analyte, Q denotes the sorption capacity and
K stands for the sorption affinity.
Slots of the class:
knum |
|
sorptionModel |
List of model parameters 'K' and 'Q'. |
knum |
Sensor number that encodes a UNIMAN
sorption profile (1:17). The default value is
1. |
gases |
Gas indices. |
ngases |
The number of gases. |
gnames
|
Names of gases. |
concUnits |
Concentration units external to the model, values given in an input concentration matrix. |
concUnitsInt
|
Concentration units internal for the model, values used numerically to evaluate the Langmuir relation. |
sorptionModel |
A list that contains the Langmuir parameters. |
srdata |
The reference
data of Lanmuir parameters from UNIMAN dataset (see
UNIMANsorption). |
alpha |
(parameter of sensor non-linearity in mixtures) A scaling
coefficient of non-linearity induced via the affinity
parameter K. The default value is 2.25. |
Methods of the class:
predict |
Predicts a model response to an input concentration matrix. |
The plot method has three types (parameter
y):
response |
(default) Shows a modeled trasnformation of concentration profile per analyte. |
data |
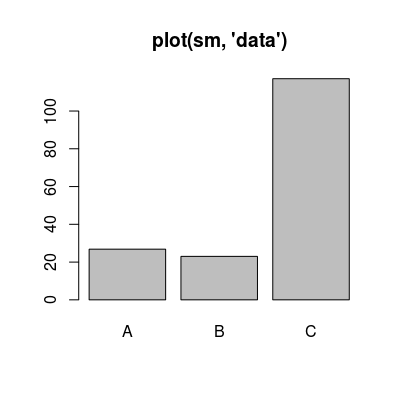
Shows the reference data from UNIMAN dataset. |
predict |
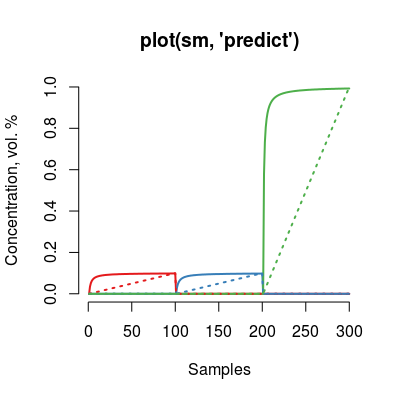
Depicts input and ouput of the model for all analytes. |
Note
We introduce a single parameter alpha of the model
to control the level of non-linearity simulated by the
Langmuir isotherm. This parameter alpha defines a
normalization across the 17 UNIMAN sorption profiles from
dataset UNIMANsorption, scaling K
values based on other two parameters Kmin (default
value 1 and Kmax (default value
150). Normalization can be disable by setting
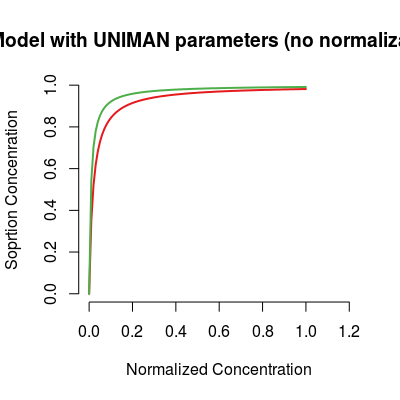
parameter Knorm to FALSE, that results in
usage of the sorption K parameters, equal to
UNIMAN ones.
Examples
# sorption model: default initialization sm <- SorptionModel() # get information about the model show(sm)Sorption Model (knum 1), alpha 2.25print(sm)Sorption Model - knum 1 - 3 gases A, B, Cplot(sm)
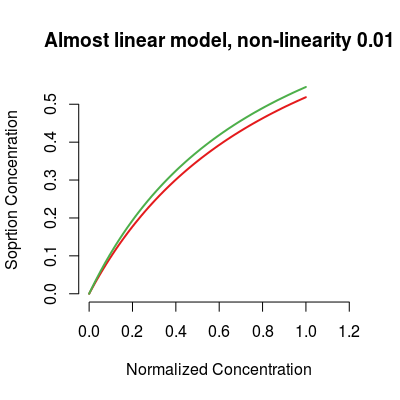
# model: custom parameters # almost linear model sm <- SorptionModel(alpha=0.01, gases=c(1, 3)) plot(sm, main="Almost linear model, non-linearity 0.01")
# non-linear model sm <- SorptionModel(alpha=0.3, gases=c(1, 3)) plot(sm, main="Non-linear model, non-linearity 0.5")
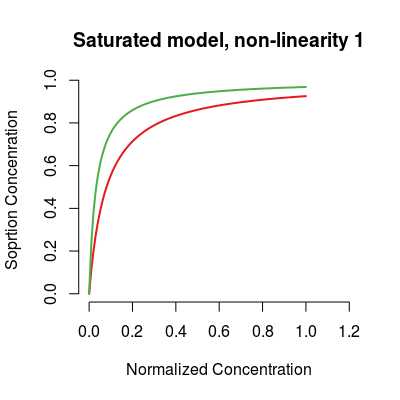
# saturated model sm <- SorptionModel(alpha=0.5, gases=c(1, 3)) plot(sm, main="Saturated model, non-linearity 1")
# model with UNIMAN sorption parameters sm <- SorptionModel(Knorm=FALSE, gases=c(1, 3)) plot(sm, main="Model with UNIMAN parameters (no normalization)")
# method plot # - plot types 'y': response, data, predict sm <- SorptionModel() # default model plot(sm, "response", main="plot(sm, 'response')")
# default plot type, i.e. 'plot(sm)' does the same plotting plot(sm, "data", main="plot(sm, 'data')")
plot(sm, "predict", main="plot(sm, 'predict')")
See also
UNIMANsorption